Cell-free DNA Fragmentomics: A Promising Predictor of Cancer
Cell-free DNA (cfDNA) represents extracellular DNA secreted into the bloodstream by cells undergoing programmed cell death. Notably, a portion of this cfDNA originates from cancer cells, termed circulating tumor DNA (ctDNA). This ctDNA holds significant potential for non-invasive early cancer detection through a simple blood draw.
A key characteristic of cfDNA is its fragmented state, resulting from DNA disassembly mechanisms during cell death. While cfDNA from healthy cells follows a somewhat predictable fragmentation pattern, cfDNA from cancer cells exhibits more disorganized and chaotic fragmentation. Recognizing and analyzing these disturbances in fragmentation can enable the prediction of cancer, even in its early stages.
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics, a field that has garnered substantial interest and holds a bright future in cancer research.
The Significance of cfDNA Fragmentation
cfDNA consists of DNA fragments in plasma derived from apoptotic and necrotic cells with different topologies. Although once considered mere cellular waste, cfDNA is now recognized as a critical biomarker reflecting the physiological state, particularly through ctDNA. The predictive value of cfDNA and ctDNA has been harnessed in various areas, such as non-invasive prenatal diagnosis, cancer detection and monitoring, and minimal residual disease (MRD) monitoring.
However, distinguishing between normal-derived cfDNA and tumor- or fetal-derived cfDNA poses significant challenges. Traditional sequence analysis is insufficient, as tumor-specific alterations are rare. Consequently, probing the fragment characteristics of cfDNA at the genome-wide level offers a more effective approach for biomarker discovery. Tumor-derived cfDNA exhibits distinct fragment characteristics compared to normal-cell-derived cfDNA, a phenomenon that fragmentomics research aims to leverage to enhance diagnostic methods (Qi et al).
cfDNA predominantly originates from blood cells, with non-random fragmentation patterns corresponding to nucleosome lengths—mononucleosome (~167 bp) and dinucleosome (~334 bp). This pattern arises because digestive enzymes can only access DNA where it is not coiled around histones, specifically at the ends of nucleosomes (Figure 1). Studies have shown that the cleavage of cfDNA by nucleases is selective, with specific end motifs and preferred end coordinates. These can reflect underlying chromatin accessibility and gene expression patterns and can indicate the tissue of origin (Snyder et al).
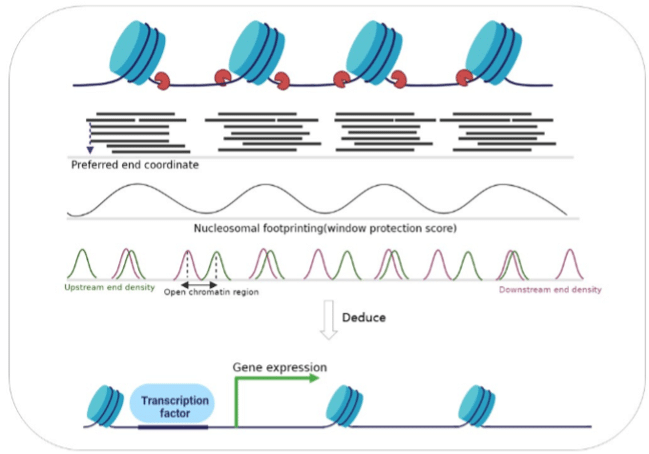
Figure 1: The uneven distribution of cfDNA fragments in the genome due to histone protection reveals tissue-specific nucleosome positioning, transcription factor binding sites, and gene expression. (From Qi et al)
The fragment size of cfDNA is a critical aspect of its analysis. ctDNA exhibits higher fragmentation, the underlying mechanism for this phenomenon may be related to the cleavage sites of nucleases. Attempts have been made to identify ctDNA through the enrichment of short molecules, thereby enhancing its applicability. Interestingly, advances in single-molecule sequencing technology have recently uncovered many long cfDNA fragments in maternal plasma (Yu et al).
The ends of cfDNA fragments also show regular characteristics, including specific end motifs, jagged ends, and preferred end coordinates. Certain genomic locations, known as plasma DNA preferred ends, are preferentially cleaved when plasma DNA molecules are formed. The preferred end distribution reflects nuclease activity, which may be distorted in disease, thereby studying these patterns may be predictive for cancer and other syndromes (Zhou et al). A class of ctDNA signature in the form of preferred end coordinates correlates with the tumor fraction in hepatocellular carcinoma (Qi et al).
Selected Research Areas
1. Cancer Early Detection with Fragmentation Patterns
Christiano et al explore the diagnostic potential of analyzing genome-wide cfDNA fragmentation patterns to detect and monitor cancer. The researchers developed the DNA evaluation of fragments for early interception (DELFI) approach, which uses low-coverage whole-genome sequencing (WGS) and intricate machine learning methods to examine cfDNA fragmentation profiles. By comparing profiles of cancer patients with healthy individuals, the study aims to identify specific patterns indicative of cancer. The authors also compare the fragmentation profiles of cancer patient groups with different cancer indications (Figure 2), showing that the fragmentation profiles also differ among different cancer types, pointing to the possibility of cancer-specific analyses.
2. Gene Expression
EPIC-seq, a deep sequencing method, enables high-resolution profiling of cfDNA fragmentation, facilitating detailed gene-level analyses. This technique enables the measurement of a metric called promoter fragmentation entropy, which quantifies the variability and disorder in cfDNA fragment ends around gene promoters and is highly correlated with gene expression. This way, researchers can infer the activity level of genes, providing a means to study gene regulation and chromatin dynamics in various biological contexts and diseases (Esfahani et al).
3. Fragmentation and Epigenetics
cfDNA fragmentation is heavily influenced by DNA methylation (Ivanov et al), and researchers have made attempts to reverse engineer the methylation patterns governing fragmentation to trace back to the original epigenetic regulation marks (Zhou et al). By using this technique, researchers may circumvent the need to perform the costly and complex methylation-specific sequencing steps.
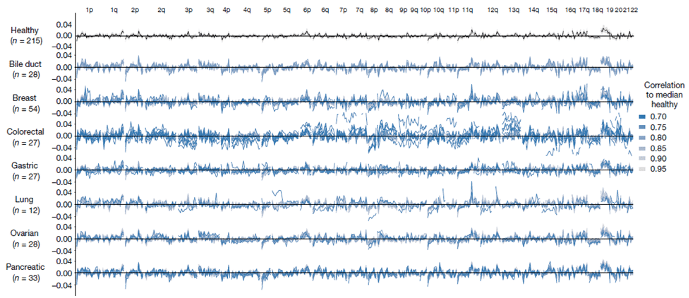
Figure 2: Genome-wide fragmentation profiles differ not only between cancer-control groups, but also between groups of patients with different cancer types. (From Christiano et al)
Conclusion
cfDNA fragmentomics represents a versatile and promising direction for early, non-invasive cancer detection. By exploiting the unique properties of cfDNA fragmentation, researchers can gain a fuller picture of disease presence and integrate this knowledge with other omics fields, such as epigenomics and transcriptomics. The future of cfDNA fragmentomics in cancer research and beyond appears exceptionally bright.
Recent blogs
A New Era in Liver Cancer Detection: The Promise of HepaAiQ
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
New developments in the field of circulating tumor cells (2024)
The blog post focuses on how researchers can produce more meaningful, applicable results that directly benefit human health.
Singlera technologies 5: Panseer – detecting pan-cancer signatures years before conventional diagnosis
In the last blog of the year and the concluding chapter of the Singlera series, we are going to explore PanSeer, a blood-based screening test utilizing unique methylation signatures.