A New Era in Liver Cancer Detection: The Promise of HepaAiQ
Early detection and prognosis evaluation for hepatocellular carcinoma by circulating tumour DNA methylation: A multicentre cohort study (DZ Guo et al., 2024)
Introduction
Liver cancer is one of the deadliest cancers worldwide, with hepatocellular carcinoma (HCC) accounting for 80% of cases. Early detection is crucial, as it offers the best chance for curative treatment, yet existing methods like ultrasound and alpha-fetoprotein (AFP) testing often fall short, especially in identifying early-stage disease. But now, a new blood-based test called HepaAiQ is poised to revolutionize how we detect and manage HCC.
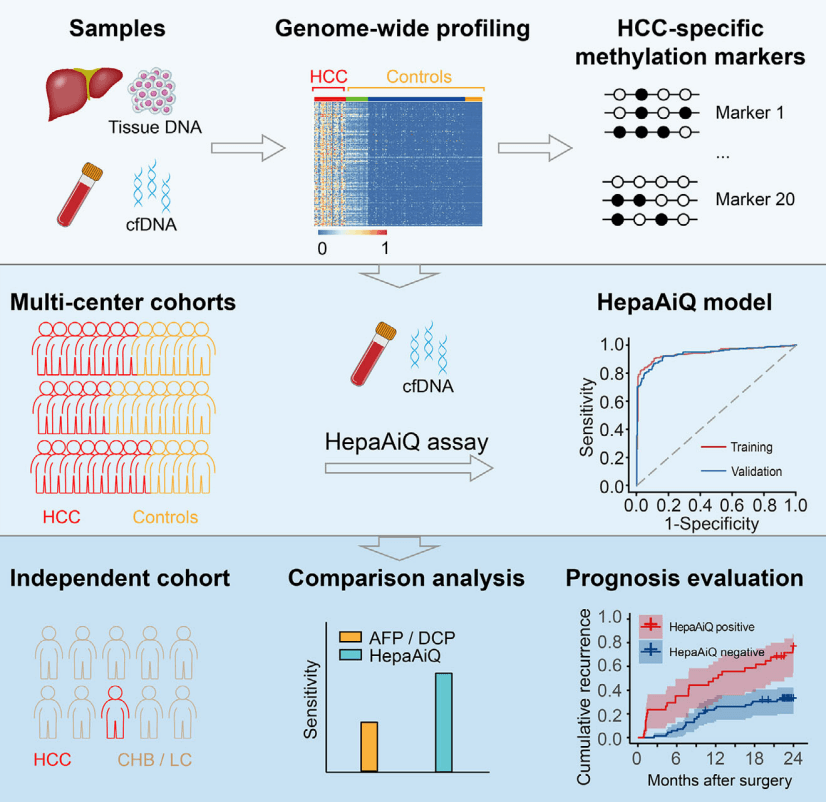
The HepaAiQ assay integrates the most effective HCC-specific methylation biomarkers using multilocus quantitative PCR. In multicentre studies, the HepaAiQ model accurately distinguishes HCC from other liver diseases, surpassing existing serum tests. The HepaAiQ excels in early-stage HCC detection in high-risk patients and postresection assessment, potentially fulfilling clinical needs with high accuracy, accessibility and affordability.
What Is HepaAiQ?
HepaAiQ is a cutting-edge diagnostic tool that leverages the power of circulating tumor DNA (ctDNA) methylation markers. ctDNA is shed by cancer cells into the bloodstream, carrying unique DNA methylation patterns that can serve as precise biomarkers for specific cancers. While previous approaches relied on complex and costly next-generation sequencing (NGS) technologies, HepaAiQ simplifies the process by using quantitative methylation-specific PCR (qMSP), making it faster, more affordable, and suitable for clinical practice.
The Science Behind the Innovation
In a multicenter study involving over 1,000 participants, HepaAiQ demonstrated remarkable accuracy in detecting HCC. Researchers screened hundreds of tumor samples and healthy controls to identify the most reliable DNA methylation markers specific to HCC. These markers were then integrated into the qMSP-based HepaAiQ model, which was validated across multiple cohorts.
The results speak for themselves:
Genome-wide methylation profiling was conducted to identify differentially methylated regions (DMRs) distinguishing HCC tumours from peritumoural tissues and healthy plasmas. The twenty most effective DMRs were verified and incorporated into a multilocus qMSP assay (HepaAiQ). The HepaAiQ model was trained to separate 293 HCC patients (Barcelona Clinic Liver Cancer (BCLC) stage 0/A, 224) from 266 controls including chronic hepatitis B (CHB) or liver cirrhosis (LC) (CHB/LC, 96), benign hepatic lesions (BHL, 23), and healthy controls (HC, 147). The model achieved an area under the curve (AUC) of 0.944 with a sensitivity of 86.0% in HCC and a specificity of 92.1% in controls. Blind validation of the HepaAiQ model in a cohort of 523 participants resulted in an AUC of 0.940 with a sensitivity of 84.4% in 205 HCC cases (BCLC stage 0/A, 167) and a specificity of 90.3% in 318 controls (CHB/LC, 100; BHL, 102; HC, 116). When evaluated in an independent test set, the HepaAiQ model exhibited a sensitivity of 70.8% in 65 HCC patients at BCLC stage 0/A and a specificity of 89.5% in 124 patients with CHB/LC. Moreover, HepaAiQ model was assessed in paired pre- and postoperative plasma samples from 103 HCC patients and correlated with 2-year patient outcomes. Patients with high postoperative HepaAiQ score showed a higher recurrence risk (Hazard ratio, 3.33, p < .001).
Outperforming Traditional Biomarkers
Unlike AFP and des-gamma-carboxy prothrombin (DCP), which often fail to detect early-stage HCC, HepaAiQ proved highly effective even in patients with small tumors (<2 cm) or those who were AFP-negative. This means fewer missed diagnoses and better outcomes for high-risk patients.
Beyond Diagnosis: Tracking Treatment and Outcomes
One of the most exciting applications of HepaAiQ is its ability to monitor tumor dynamics post-surgery. The study revealed that ctDNA methylation levels dropped significantly after successful tumor resection. Importantly, patients with persistent high levels were more likely to experience recurrence, underscoring HepaAiQ’s potential as a prognostic tool.
A Cost-Effective Solution for Population-Level Screening
By adopting a qMSP-based approach, HepaAiQ makes advanced methylation analysis accessible to more patients. This positions it as a practical option for population-level HCC screening, particularly in high-risk groups such as those with chronic hepatitis B or liver cirrhosis. Simulation studies suggest that HepaAiQ could dramatically increase the detection rates of early-stage HCC compared to the current standard of care, potentially saving thousands of lives.
Looking Ahead
The HepaAiQ test marks a significant step forward in the fight against liver cancer. Its ability to accurately detect early-stage HCC, predict recurrence, and reduce the barriers of cost and complexity could make it a game-changer in liver cancer management. While further validation in diverse populations is needed, the future of liver cancer detection and monitoring looks brighter than ever.
For patients, clinicians, and researchers alike, HepaAiQ offers hope—a chance to detect liver cancer earlier, treat it more effectively, and improve survival outcomes.
Recent blogs
Cell-free DNA Fragmentomics: A Promising Predictor of Cancer
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
New developments in the field of circulating tumor cells (2024)
The blog post focuses on how researchers can produce more meaningful, applicable results that directly benefit human health.
Singlera technologies 5: Panseer – detecting pan-cancer signatures years before conventional diagnosis
In the last blog of the year and the concluding chapter of the Singlera series, we are going to explore PanSeer, a blood-based screening test utilizing unique methylation signatures.