Singlera Technologies 4: Catching Pancreatic Cancer early with intricate methylation markers
Highlights
One significant hurdle in PDAC diagnosis is the lack of precise early detection methods. Symptoms, including abdominal pain, weight loss, and jaundice, are nonspecific and often attributed to other gastrointestinal issues. Additionally, the widely used marker, CA19-9, while helpful, presents limitations due to its low sensitivity in early-stage disease and false positives in certain populations. Enter PDACatch, a novel assay developed to detect circulating tumor DNA methylation signatures with greater sensitivity and accuracy.
Background
Pancreatic ductal adenocarcinoma (PDAC) stands as a formidable challenge in oncology due to its aggressive nature and late-stage diagnosis, contributing to its low survival rates. One of the major hurdles in managing PDAC lies in the lack of effective early detection methods. The disease often remains asymptomatic until it reaches advanced stages, rendering treatment options limited and survival rates drastically low. The pressing need for reliable early detection methodologies has steered researchers toward exploring novel avenues in biomarker discovery.
In recent years, while researchers were looking for improved diagnostic tools, the focus has shifted towards the field of epigenetics, particularly DNA methylation. Alterations in DNA methylation patterns have been increasingly recognized as hallmarks of cancer development and progression. In PDAC, these epigenetic modifications offer a promising avenue for identifying specific biomarkers that could enable early detection, revolutionizing the clinical landscape.
Results
The research conducted to address the deficiencies in early PDAC detection led to the development of PDACatch, an innovative assay leveraging DNA methylation signatures in blood samples. The study employed Reduced Representation Bisulfite Sequencing (RRBS) to scrutinize methylation patterns across 46 PDAC tumors, 28 para-tumor tissues, and 143 plasma samples (20 PDAC, 123 healthy). The investigation focused on identifying Methylation Haplotype Blocks (MHBs) as potential markers for PDAC, distinct genomic regions characterized by tightly coupled CpG methylation sites. The definition of MHBs and their application were described more in-depth in the first blog of the series.
MHB-specific metrics were utilized to quantify methylation levels and discern MHBs containing PDAC-specific methylation haplotypes. A set of 171 de novo markers emerged from these analyses, showcasing promise for downstream investigations. To streamline marker selection, 750 markers were resulted from combining the PDAC de novo markers and existing ones from the marker panel of the pan-cancer diagnostic approach, PanSeer, another Singlera property, which will be better discussed in next month’s upcoming blog post.
The PDACatch classifier was constructed utilizing 56 of the most discriminatory markers of this combined marker panel. In the training set, this classifier exhibited a high AUC of 0.93 (sensitivity = 71%, specificity = 91%), reaffirming its robustness. Importantly, this performance was maintained in the validation set, suggesting consistency and reliability across datasets. The classifier also showed independence from demographic and clinical variables, bolstering its potential clinical utility. PDACatch showcased superior sensitivity, particularly in early-stage PDAC detection, surpassing the performance of the commonly used serum marker, carbohydrate antigen 19-9 (CA19-9). Notably, PDACatch demonstrated higher sensitivity for Stage I PDAC (sensitivity was 80% and 68% for PDACatch and CA19-9, respectively, at the same 89% specificity) and efficiently identified CA19-9-negative PDAC cases (sensitivity = 54-75%) while maintaining high specificity (91%), underscoring its value in capturing elusive early-stage PDAC cases that might evade conventional detection methods.
The point of PDACatch is not to suppress the use of CA19-9, but instead to complement its utility and serve as an effective adjuvant diagnostic method. The authors were exactly able to perceive potential synergistic benefits when merging both approaches into a combined classifier. The classifier built from CA19-9 with PDACatch presented amplified accuracy (AUC = 0.96, sensitivity = 94%, specificity = 93%), particularly in detecting early-stage PDAC, compared to individual classifiers, amplifying its potential clinical utility (Figure 1). In an independent blinded test on a separate cohort, PDACatch displayed consistent accuracy (AUC = 0.91, sensitivity = 82%, specificity = 88%) akin to the validation set, reinforcing its robustness in classifying PDAC plasma from healthy controls.
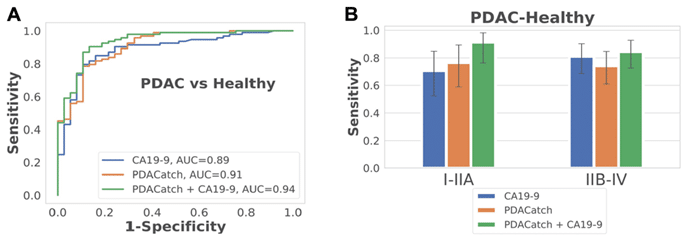
Figure 1.: PDACatch shows heightened sensitivity and specificity in separating pancreatic cancer from healthy controls in the training and validation sets, even in early stage disease, particularly when utilized in tandem with traditional blood biomarker, CA19-9.
However, certain limitations of the present paper need to be addressed in further research. Limitations include sample size constraints in specific subsets (e.g., early-stage PDAC), warranting validation in larger cohorts. Further optimization and validation in diverse populations are imperative before clinical implementation.
Conclusion
The inception of PDACatch marks a pivotal advancement in the pursuit of more effective early detection methods for PDAC. With its ability to decipher subtle methylation markers in blood samples, PDACatch has the potential to offer noninvasive and more accurate pancreatic cancer diagnosis and prognosis. As researchers continue to refine and validate this innovative approach, the promise of detecting PDAC in its infancy holds the potential to significantly impact patient outcomes and survival rates.
Recent blogs
A New Era in Liver Cancer Detection: The Promise of HepaAiQ
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
Cell-free DNA Fragmentomics: A Promising Predictor of Cancer
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
New developments in the field of circulating tumor cells (2024)
The blog post focuses on how researchers can produce more meaningful, applicable results that directly benefit human health.