Singlera technologies 3: Methylation signatures for hepatocellular carcinoma detection
Identification of DNA methylation signatures for hepatocellular carcinoma detection and microvascular invasion prediction. – Hao et al.
Highlights
Hepatocellular carcinoma (HCC) is a major contributor to cancer-related mortality worldwide and is associated with a high risk of postoperative recurrence, with microvascular invasion (MVI) being a significant factor. DNA methylation patterns have been explored in HCC research for diagnosis and prediction, but consistent markers for preoperative prediction of MVI have been lacking.
In this study, Hao et al examines DNA methylation in circulating tumor DNA (ctDNA) and establishes a highly accurate diagnostic tool for HCC, enabling reliable differentiation of HCC patients from healthy individuals non-invasively. This breakthrough offers potential for early HCC detection and improved patient outcomes. The study also identifies DNA methylation markers that can predict MVI status facilitating tailored treatments and influencing surgical strategies, particularly important for HCC patients with a high risk of early recurrence.
Introduction
In this next part of the Singlera series, we are going to look at the science of how non-invasive, methylation-based predictive testing can help alleviate the burden of hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) is a formidable adversary on the global cancer landscape, ranking as the third leading cause of cancer-related mortality. Its aggressive nature and propensity for early postoperative recurrence pose immense challenges for clinicians and patients alike. One pivotal risk factor for such recurrence is microvascular invasion (MVI), which indicates the presence of tumor emboli in vascular spaces lined by endothelial cells. Understanding and predicting MVI status is crucial for making informed surgical decisions and tailoring postoperative treatments, enhancing their efficiency.
Previous efforts in HCC research have explored the potential of DNA methylation patterns as markers for both diagnosis and prediction. DNA methylation, a well-known mechanism of epigenetic regulation, plays a crucial role in tumorigenesis and early metastasis in various cancers, including HCC. Studies have hinted at the potential of DNA methylation markers as predictive markers for HCC early detection or disease progression (Kisiel et al), but consistent and effective markers for preoperative prediction of MVI status remained elusive.
ctDNA Methylation Signature to Distinguish HCC Patients from Healthy Individuals
The authors first used samples of liver cancer and matched healthy tissue to compare their methylation profiles to select differentially methylated regions which could be utilized as specific disease markers. Genome-wide methylation sequencing was performed to observe the widest array of methylated sites possible. With this method, 65 regions, which were significantly different between normal liver tissues and HCC tissues, were found. Using these methylation markers as independent variables with supervised machine learning, they built a binary predictive model to classify normal or HCC liver tissues. The results were more than promising, with an area under the curve (AUC) score no less than 0.98.
Afterwards, they could apply this trained model on liquid biopsy samples to classify healthy and disease. The ctDNA methylation signature they uncovered exhibited remarkable accuracy, with an impressive AUC of 0.96 (Figure A). This signifies that HCC patients can now be reliably distinguished from their healthy counterparts, and such a high level of accuracy can significantly reduce the risk of misdiagnosis. As such, by harnessing the power of DNA methylation patterns in ctDNA, this technique promises to enhance patient care but also offers a cost-effective and convenient alternative to traditional diagnostic methods.
DNA Methylation Markers for the Prediction of MVI Status
Beyond predicting the existence of cancer, the study also aims to tackle microvascular invasion (MVI) status in HCC, which is a critical determinant for treatment planning and prognosis, and this study makes headway in its accurate preoperative prediction. By contrasting tissue samples of MVI-negative (MVI-) and MVI-positive (MVI+) the research team could identify specific DNA methylation markers, and with those, achieved a high level of accuracy. Binary classification based on these markers demonstrated an AUC of more than 0.85 (Figure B), firmly distinguishing MVI from MVI + HCC tissues.
The importance of MVI determination was further reinforced as the data collected for this study showed a profound difference in recurrence-free survival (RFS) of MVI groups, with MVI- patients having significantly higher likelihood of better prognosis with HCC (Figure C)
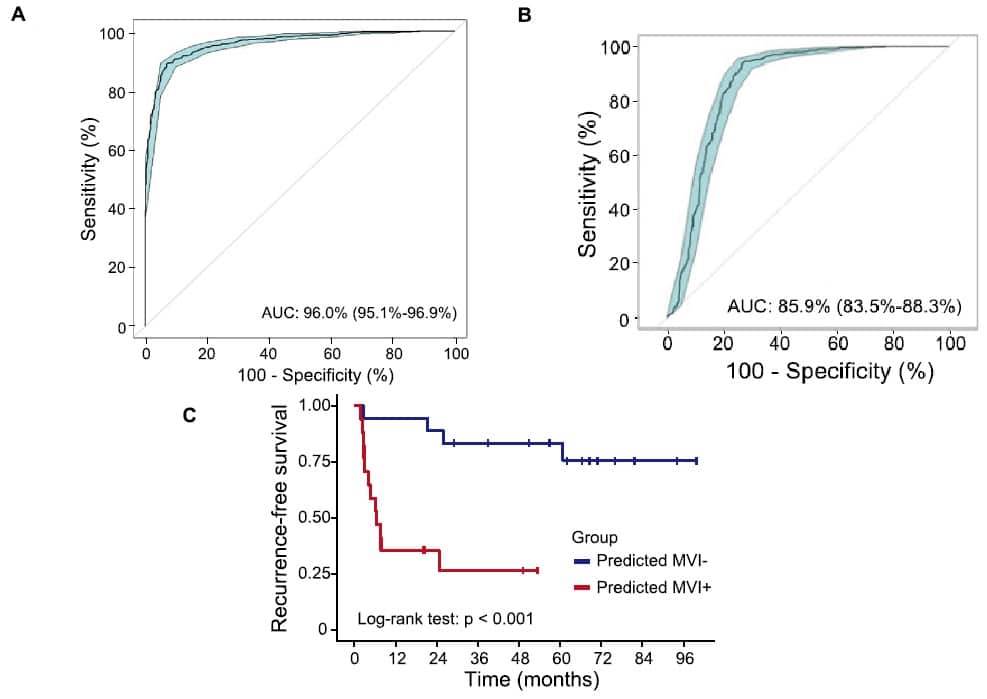
Figure ~ A: The classification model can separate normal and HCC plasma samples sensitively and specifically using the 65 MHB markers; B: MVI markers accurately classified MVI- and MVI + tissues; C: Chances for recurrence-free survival is significantly larger by the MVI- group than the MVI+ group.
Conclusion
The study harnesses the power of DNA methylation to redefine the landscape of hepatocarcinoma detection. By introducing a methylation signature of the non-invasively collectible ctDNA for early detection of cancer and tissue DNA methylation markers for MVI prediction, this research opens new doors for precision medicine in the battle against HCC. These markers show potential to enhance patient care and streamline treatment strategies against this aggressive disease.
Recent blogs
A New Era in Liver Cancer Detection: The Promise of HepaAiQ
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
Cell-free DNA Fragmentomics: A Promising Predictor of Cancer
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
New developments in the field of circulating tumor cells (2024)
The blog post focuses on how researchers can produce more meaningful, applicable results that directly benefit human health.