Singlera technologies 2: Early detection of residual molecular disease in colorectal cancer through circulating tumor DNA methylation
Colorectal cancer (CRC), the second most common cause of cancer-related death, has a 5-year survival rate as low as 60%. This is partly due to the relatively high recurrence that can be experienced in patients after radical surgery. Moreover, one of the most critical issues in the management of CRC is the selection of patients for adjuvant chemotherapy after surgery, which should be based on risk stratification. This decision is currently determined by the CRC stage and clinical risk factors.
For these reasons, there is an urgent need for an effective tool for early detection of disease recurrence which may also improve the treatment of patients with cancer.
This study sought to answer the question of whether longitudinal changes in ctDNA methylation are effective in monitoring disease progression from molecular residual disease to recurrence.
Therefore, the objective of the study was to assess ctDNA using 6 DNA methylation markers in blood samples and to evaluate the association between the presence of ctDNA and CRC recurrence during the course of the disease.
S Mo et al. performed a multicenter prospective longitudinal cohort study of 299 patients with stage I to III CRC. The study was conducted from December 12, 2019, to February 28, 2022, and the blood samples were collected from 2 hospitals before and after surgery, during and after adjuvant chemotherapy, and every 3 months for up to 2 years.
The study utilized a simplified multiplex quantitative polymerase chain reaction method to analyze changes in ctDNA methylation from plasma samples. The status of ctDNA methylation was determined using the ColonAiQ assay, which measured the methylation status of 6 specific genes. The presence of ctDNA was defined as having at least 1 positive marker, while the absence of ctDNA was defined as ctDNA negative. The 6 ctDNA methylation markers were the following BCAN which indicates brevican; BCAT1, branched-chain amino acid transaminase 1; IKZF1, ikaros family zinc finger 1; Septin9_1, septin 9 locus 1; Septin9_2, septin 9 locus 2; and VAV3, vav guanine nucleotide exchange factor 3.
To assess the residual molecular tumor after surgery and to stratify the risk of recurrence in patients, ctDNA was tested in plasma samples collected approximately 1 month after surgery. Researchers found that ctDNA analysis at postoperative month 1 can effectively detect CRC recurrence. ctDNA-positive patients at this point had a significantly higher risk (17.5 times more likely to relapse) of recurrence than those who tested negative.
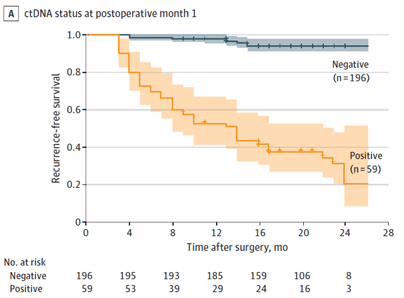
Figure A.: Kaplan-Meier estimates of recurrence-free survival, stratified by ctDNA status at postoperative month 1.
At the end of follow-up, 94,4% of ctDNA-negative patients were relapse-free, while 66,1% of ctDNA-positive patients had recurrence. The sensitivity for relapse detection, at postoperative month 1, was 78,0% and the specificity was 90,2%. This suggests that ctDNA analysis at postoperative month 1 can be a reliable method for detecting molecular residual disease and predicting recurrence in CRC patients.
Researchers also tried to establish an association between methylation markers and recurrence, therefore they examined the positive rates of 6 markers individually. However, the conclusion was that the combination of the 6 markers captures a fuller spectrum of recurrence than individual markers alone.
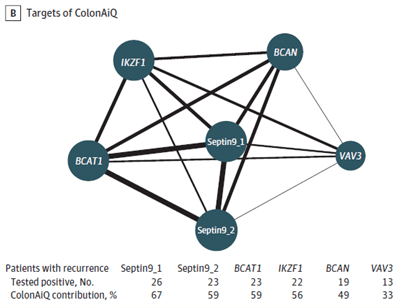
Figure B.: Network map of 6 targets of the ColonAiQ test. The size of the nodes is proportional to the number of patients who tested positive, and the thickness of the lines is proportional to the number of patients who tested positive for 2 targets and had a recurrence.
To improve the detection of relapse, researchers combined the status of ctDNA and carcinoembryonic antigen (CEA) – that are routinely used in clinics – at postoperative month 1. The integration of ctDNA and CEA reached an area under the curve of 0.849, while ctDNA and CEA alone were 0.839 and 0.623, respectively. Patients with positive test results in either ctDNA or CEA had inferior prognoses compared to those with negative results. The combined test detected recurrence with 83.3% sensitivity and 86.5% specificity. Overall, the combination has shown improved efficiency in detecting recurrence in patients with CRC.
The study suggests that postoperative month 1 ctDNA can potentially guide the choice of adjuvant treatment for patients with stage III CRC. Patients with stage III CRC who had a negative ctDNA test at postoperative month 1 had a significantly better outcome, regardless of clinical risk classification. On the other hand, ctDNA-positive patients had a higher risk of recurrence. Thus, ctDNA status at postoperative month 1 was strongly associated with prognosis in patients treated with different durations and intensities of adjuvant chemotherapy, suggesting that ctDNA status performed better in discriminating prognosis than current classifications.
S Mo et al. also evaluated the presence of ctDNA after the completion of adjuvant chemotherapy from the first blood sample collected after treatment. Of the 149 patients with blood samples obtained after adjuvant chemotherapy, 22 experienced recurrence. 12 of the 18 ctDNA-positive patients (66.7%) had relapsed, and 121 of the 131 ctDNA-negative patients (92.4%) remained recurrence-free. Patients who tested positive for ctDNA had a significantly shorter recurrence-free survival compared to those who tested negative. These results suggest that the presence of ctDNA after adjuvant chemotherapy showed risk discrimination in recurrence-free survival, indicating the potential of ctDNA analysis in predicting disease recurrence.
The longitudinal analysis of ctDNA after definitive treatment showed a discriminating effect in predicting recurrence risk in patients with CRC. Patients who tested positive for ctDNA had a significantly shorter recurrence-free survival compared to those who tested negative.
The persistent ctDNA-positive patients were 68.8 times more likely to have a recurrence than persistent ctDNA-negative patients, in this study only 4 of 133 patients (3%) with serially negative ctDNA relapsed.
Therefore, the presence of ctDNA after curative treatment was associated with a higher risk of recurrence, moreover, ctDNA was detected earlier than radiologically confirmed recurrence – with a median lead time of 3,3 months – indicating its potential as an early detection tool.
In conclusion, the cohort study proved the effectiveness of ctDNA methylation in monitoring disease progression from molecular residual disease to recurrence in CRC patients. The presence of ctDNA was strongly associated with recurrence before and after surgery, after adjuvant chemotherapy, and during longitudinal monitoring. The results suggest a potential role for ctDNA methylation in postoperative management and may serve as a pragmatic tool for residual disease detection, risk stratification, chemotherapy guidance, and clinical monitoring of recurrence.
Reference
Mo S, Ye L, Wang D, Han L, Zhou S, Wang H, Dai W, Wang Y, Luo W, Wang R, Xu Y, Cai S, Liu R, Wang Z, Cai G. Early Detection of Molecular Residual Disease and Risk Stratification for Stage I to III Colorectal Cancer via Circulating Tumor DNA Methylation. JAMA Oncol. 2023 Jun 1;9(6):770-778. doi: 10.1001/jamaoncol.2023.0425. PMID: 37079312; PMCID: PMC10119774.
Recent blogs
A New Era in Liver Cancer Detection: The Promise of HepaAiQ
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
Cell-free DNA Fragmentomics: A Promising Predictor of Cancer
In this blog entry, we will explore the recent history, intriguing findings, and tools related to cfDNA fragmentomics.
New developments in the field of circulating tumor cells (2024)
The blog post focuses on how researchers can produce more meaningful, applicable results that directly benefit human health.